My lab focuses on statistical modeling of neural signals at different spatial scales: spikes, LFP, ECoG, and EEG/MEG. Propagation of electrical and magnetic fields in the brain depends on both static anatomical features and state-dependent dynamical features like coherence, neuromodulation, and active pathways. I’m particularly interested in constructing models that use strong prior knowledge about these features in specific brain states to bridge spatial scales. Such models could explain apparent discrepancies in signals recorded at different scales and improve our understanding of the micro-scale dynamics that underlie macro-scale biomarkers of pathological states.
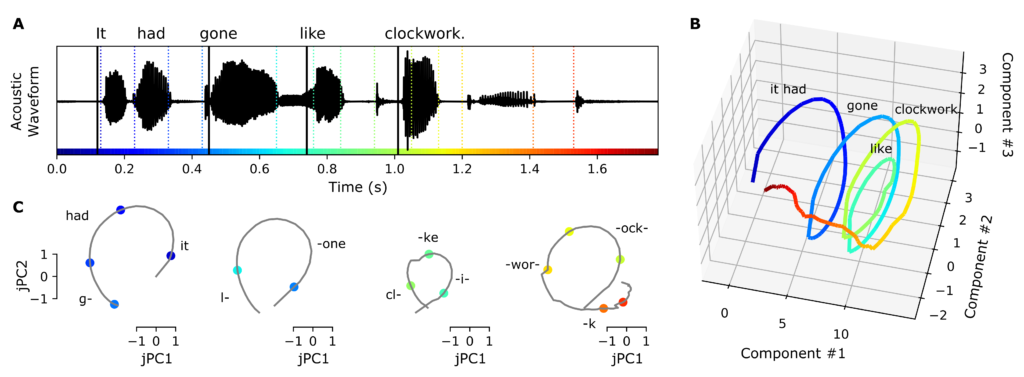 From 2019 until 2021 I worked as a postdoctoral scholar in the Chang Lab at UCSF, using statistical modeling techniques to study the cross-scale neural basis of speech perception using ECoG recordings. While it has been known for some time that phonetic features and some timing cues are dynamically encoded in specific brain areas, there is controversy regarding where and how these low-level features are grouped into longer chunks such as words and phrases. Using a group-reduced-rank regression model, I showed that timing-related neural activity over the superior temporal gyrus during speech perception supports a low-dimensional latent state representation that is distributed over centimeters of cortex. The spiral-shaped dynamics of the timing representations could serve to provide temporal context for, and possibly bind across time, the concurrent processing of individual phonetic features, to compose higher-order (e.g. word-level) representations. This work is published in the journal Hearing Research.
From 2019 until 2021 I worked as a postdoctoral scholar in the Chang Lab at UCSF, using statistical modeling techniques to study the cross-scale neural basis of speech perception using ECoG recordings. While it has been known for some time that phonetic features and some timing cues are dynamically encoded in specific brain areas, there is controversy regarding where and how these low-level features are grouped into longer chunks such as words and phrases. Using a group-reduced-rank regression model, I showed that timing-related neural activity over the superior temporal gyrus during speech perception supports a low-dimensional latent state representation that is distributed over centimeters of cortex. The spiral-shaped dynamics of the timing representations could serve to provide temporal context for, and possibly bind across time, the concurrent processing of individual phonetic features, to compose higher-order (e.g. word-level) representations. This work is published in the journal Hearing Research.
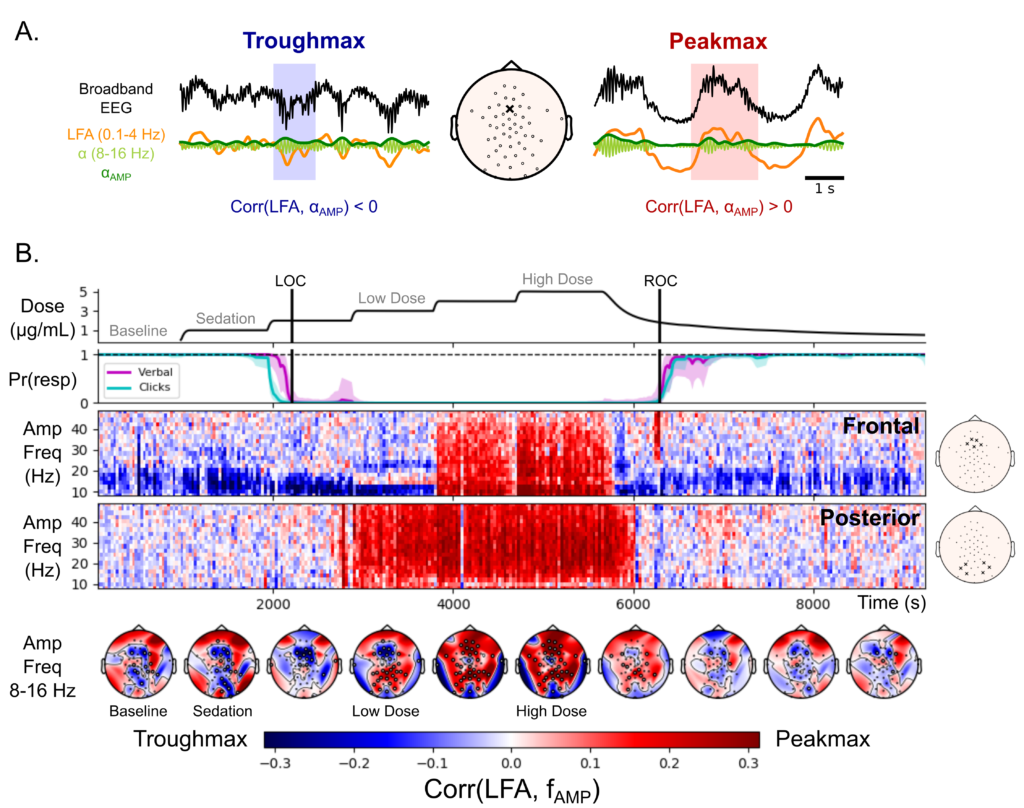 From 2015 until 2019 I worked with Drs. Emery Brown and Patrick Purdon at MIT and MGH, focusing on brain rhythms in anesthesia. Specifically, I used source localization techniques to identify brain regions involved in generating anesthesia-related rhythmic activity recorded from high-density EEG. In the process, I found a new marker of depth of propofol anesthesia: the coupling between broadband activity and the slow wave occurs in different locations during distinct states of unconsciousness. This observation allowed us to make inferences about the brain functions that are disrupted in progressively deeper states of anesthetic-induced unconsciousness, reported here.
From 2015 until 2019 I worked with Drs. Emery Brown and Patrick Purdon at MIT and MGH, focusing on brain rhythms in anesthesia. Specifically, I used source localization techniques to identify brain regions involved in generating anesthesia-related rhythmic activity recorded from high-density EEG. In the process, I found a new marker of depth of propofol anesthesia: the coupling between broadband activity and the slow wave occurs in different locations during distinct states of unconsciousness. This observation allowed us to make inferences about the brain functions that are disrupted in progressively deeper states of anesthetic-induced unconsciousness, reported here.
For my Ph.D. thesis “Characterizing dynamically evolving functional networks in humans with application to speech” (2015), I worked on developing methods to infer dynamic functional connectivity in brain voltage data during a task (PDF). This work was performed in the Boston University Graduate Program for Neuroscience in the lab of Dr. Frank Guenther and in collaboration with Drs. Uri Eden and Mark Kramer.
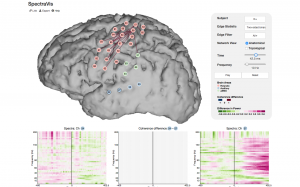
Eric Denovellis and I developed an interactive visualization (a static snapshot is shown on the right) displaying networks inferred from ECoG data during an overt reading task: SpectraVis.